Discrete Binning Effects¶
When computing the power spectrum from galaxy survey data, we estimate
the power in bandpowers, averaging over a finite set of modes in each
\(k\) or \(\mu\) bin. Especially on large scales (low \(k\)),
where we are averaging over relatively few values, this averaging
procedure can lead to additional effects on the power spectrum. The
pyRSD package accounts for these effects by applying an additional transfer
function to the continuous power spectrum that is returned by
GalaxySpectrum.power() or GalaxySpectrum.poles().
The discretely-binned power spectrum theory is computed by either the
PkmuTransfer or PolesTransfer classes, depending if the user
wants \(P(k,\mu)\) or \(P_\ell(k)\). A transfer function can be
initialized, and then the discretely-binned power spectrum theory will be
returned by the GalaxySpectrum.from_transfer() function.
Specifying the (\(k\), \(\mu\)) Grid¶
In order to account for the discrete binning effects, the user must supply the average \(k\) and \(\mu\) values on the 2D binning grid, as well as the number of modes averaged over in each bin. These bins should be finely spaced, typically about ~100 \(\mu\) bins yields good results. The continuous theory will be evaluted for each these discrete bins, and then averaged over, weighted by the number of modes in each bin, to account for any binning effects.
The class that handles setting up the \(P(k,\mu)\) grid is
PkmuGrid. This class can be initialized
# set up fake 1D k, mu bins
In [1]: k_1d = numpy.arange(0.01, 0.4, 0.005)
In [2]: mu_1d = numpy.linspace(0, 1.0, 100)
# convert to a 2D grid
# shape is (78, 100)
In [3]: k, mu = numpy.meshgrid(k_1d, mu_1d, indexing='ij')
# assign random weights for each bin for illustration purposes
In [4]: modes = numpy.random.random(size=k.shape)
# simulate missing data
In [5]: missing = numpy.random.randint(0, numpy.prod(modes.shape), size=10)
In [6]: modes.flat[missing] = numpy.nan
# initialize the grid
In [7]: grid = PkmuGrid([k_1d, mu_1d], k, mu, modes)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
<ipython-input-7-235933c43c91> in <module>()
----> 1 grid = PkmuGrid([k_1d, mu_1d], k, mu, modes)
NameError: name 'PkmuGrid' is not defined
In [8]: print(grid)
������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������---------------------------------------------------------------------------
NameError Traceback (most recent call last)
<ipython-input-8-8e1381097b77> in <module>()
----> 1 print(grid)
NameError: name 'grid' is not defined
Once a PkmuGrid object has been initialized, we can save it to disk
as a plaintext file for later use.
# save to a plaintext file
In [9]: grid.to_plaintext('pkmu_grid.dat')
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
<ipython-input-9-f4b63e7a5632> in <module>()
----> 1 grid.to_plaintext('pkmu_grid.dat')
NameError: name 'grid' is not defined
# read the grid back from disk
In [10]: grid_2 = PkmuGrid.from_plaintext('pkmu_grid.dat')
������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������---------------------------------------------------------------------------
NameError Traceback (most recent call last)
<ipython-input-10-7de7eb7ca8d9> in <module>()
----> 1 grid_2 = PkmuGrid.from_plaintext('pkmu_grid.dat')
NameError: name 'PkmuGrid' is not defined
In [11]: print(grid_2)
����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������---------------------------------------------------------------------------
NameError Traceback (most recent call last)
<ipython-input-11-fb6d86bbdf3d> in <module>()
----> 1 print(grid_2)
NameError: name 'grid_2' is not defined
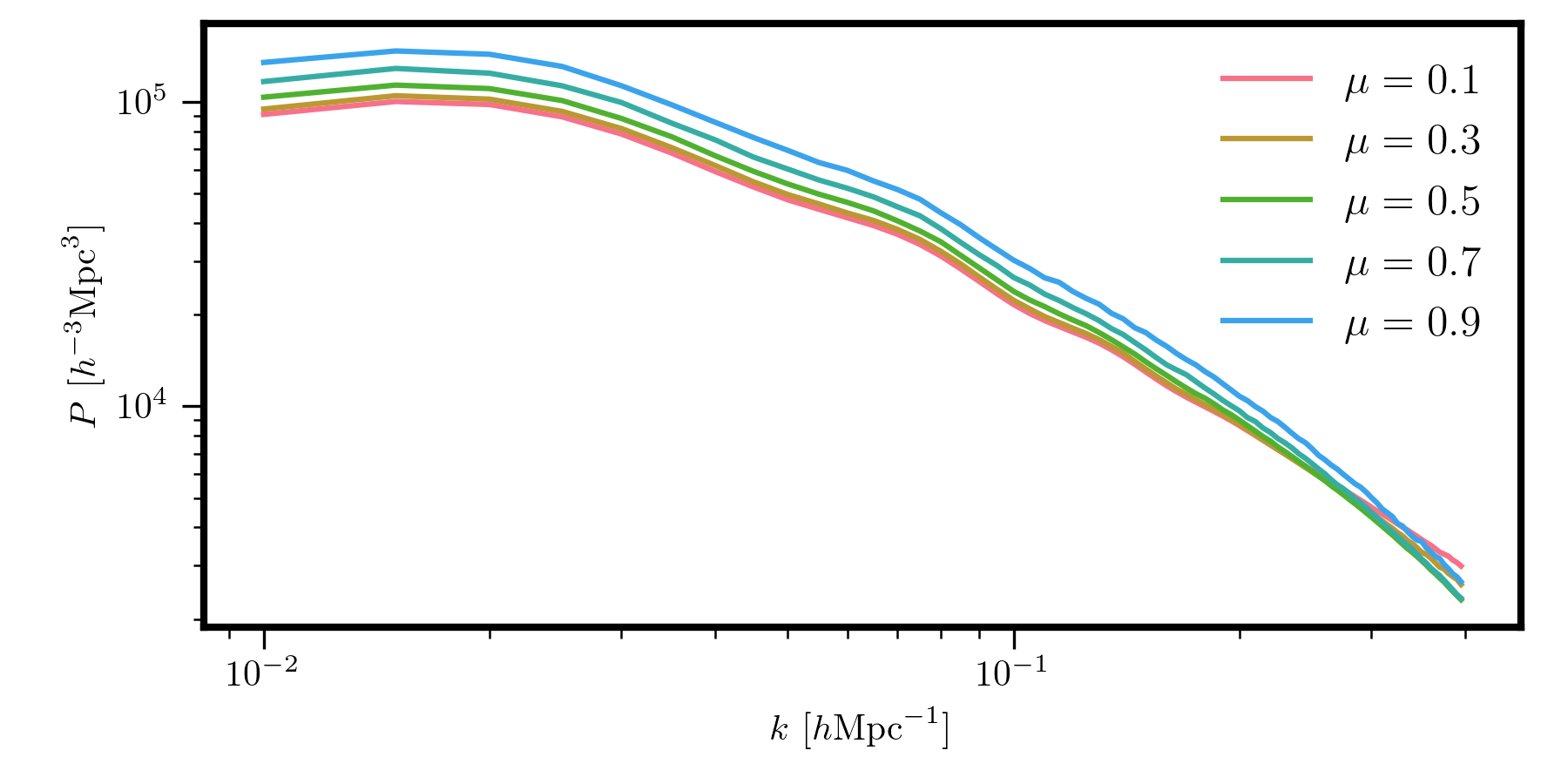
Discrete \(P(k,\mu)\)¶
Once the binning grid is initialized, we next must initialize the desired transfer function. Imagine we want to compute the power spectrum in 5 \(\mu\) bins, accounting for discrete binning effects. This can be achieved as
# edges of the mu bins
mu_bounds = [(0., 0.2), (0.2, 0.4), (0.4, 0.6), (0.6, 0.8), (0.8, 1.0)]
# the transfer function, with specified valid k range
transfer = PkmuTransfer(grid, mu_bounds, kmin=0.01, kmax=0.4)
# evaluate the model with this transfer function
Pkmu_binned = model.from_transfer(transfer)
# get the coordinate arrays from the grid
k, mu = transfer.coords # this has shape of (Nk, Nmu)
for i in range(mu.shape[1]):
plt.loglog(k[:,i], Pkmu_binned[:,i], label=r"$\mu = %.1f$" % mu[:,i].mean())
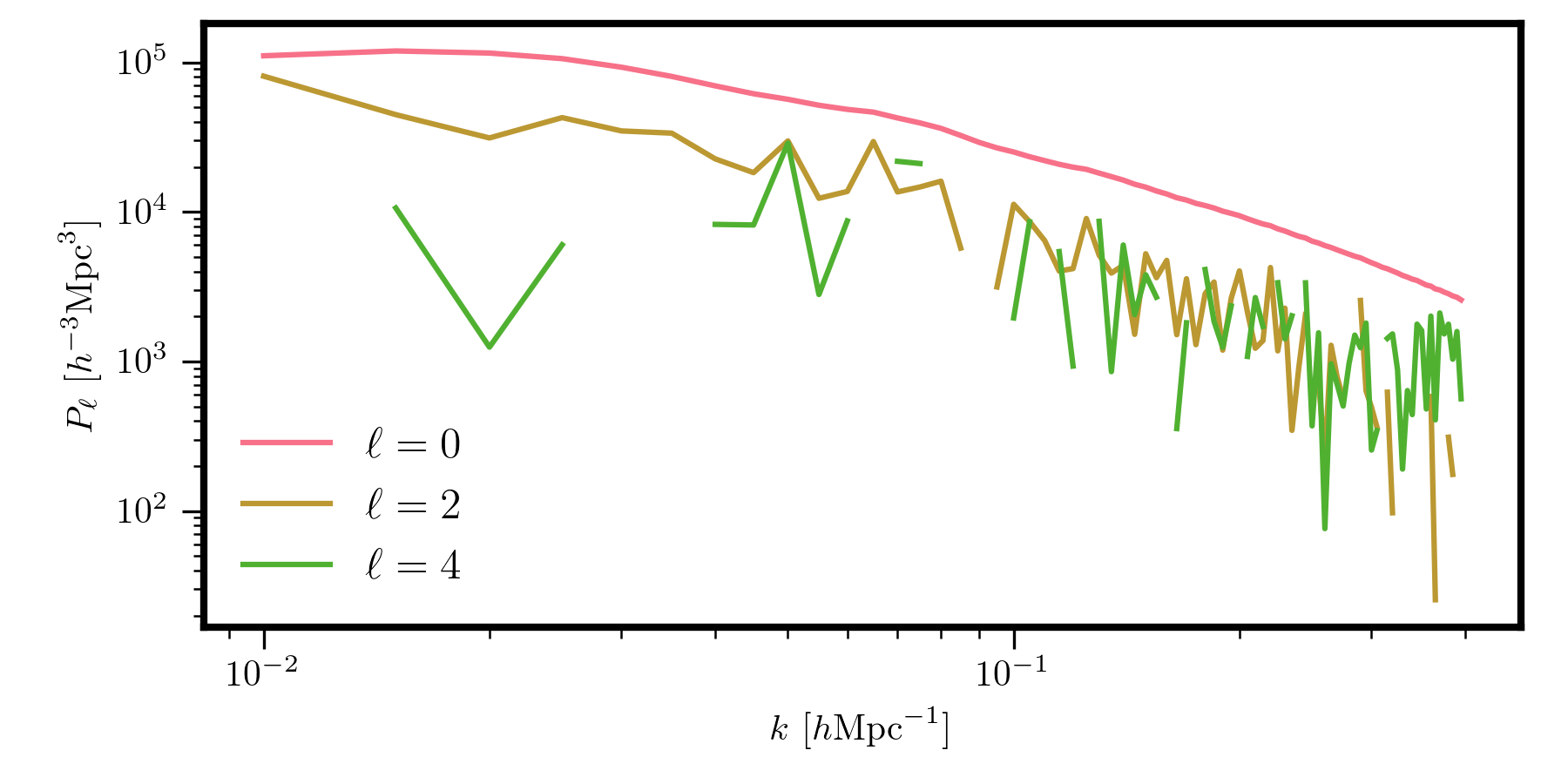
Discrete \(P_\ell(k)\)¶
We can similarly compute multipoles while accounting for discrete binning effects. This can be achieved as
# the multipoles to compute
ells = [0, 2, 4]
# the transfer function, with specified valid k range
transfer = PolesTransfer(grid, ells, kmin=0.01, kmax=0.4)
# evaluate the model with this transfer function
poles_binned = model.from_transfer(transfer) # shape is (78, 3)
# get the coordinate arrays from the grid
k, mu = transfer.coords # this has shape of (Nk, Nmu)
for i, iell in enumerate(ells):
plt.loglog(k[:,i], poles_binned[:,i], label=r"$\ell = %d$" % iell)